

This is an embedded Microsoft Office presentation, powered by Office Online. You can DOWNLOADthe PPT by clicking on the download link below the preview… Learn more: Notes on 4 Levels of Protein Structure

Potential in tasks requiring many predictions.Structure of Proteins PPT : The present PPT on Protein Structure Discuees the follwoing topics – Protein Structure, Four Levels of Protein Structure, Primary Structure of Protein, Secondary Structure of Protein, Tertiary Structure of Proteins, Quaternary Structure of Proteins, Bonds Involved in Protein Structures, Peptide Bond, Hydrogen Bond, Hydrophobic Interactions, Hydrophilic Interactions, Alpha Helix, Beta Plats, Beta Conformations, Domains and Motifs. The mainstream pipelines for protein structure prediction, demonstrating its Furthermore, HelixFold-Single consumes much less time than HelixFold-Single is validated in datasets CASP14 and CAMEO, achievingĬompetitive accuracy with the MSA-based methods on the targets with large Model to predict the 3D coordinates of atoms from only the primary sequence. Then, by combining the pre-trained PLM and theĮssential components of AlphaFold2, we obtain an end-to-end differentiable Learning paradigm, which will be used as an alternative to MSAs for learning With thousands of millions of primary sequences utilizing the self-supervised HelixFold-Single, first pre-trains a large-scale protein language model (PLM) Geometric learning capability of AlphaFold2. Proposed to combine a large-scale protein language model with the superior Prediction by using only primary sequences of proteins. Protein databases is time-consuming, usually taking dozens of minutes.Ĭonsequently, we attempt to explore the limits of fast protein structure
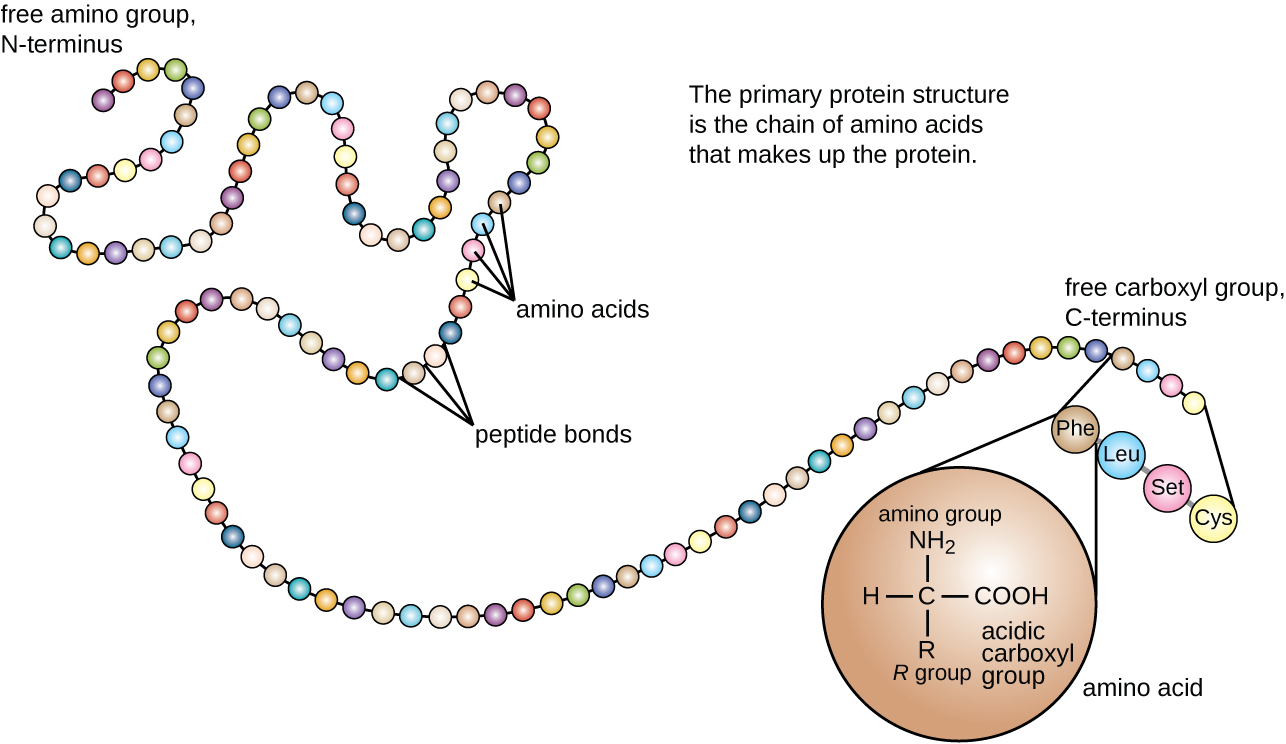
Information from the homologous sequences. Multiple Sequence Alignments (MSAs) as inputs to learn the co-evolution Download a PDF of the paper titled HelixFold-Single: MSA-free Protein Structure Prediction by Using Protein Language Model as an Alternative, by Xiaomin Fang and 9 other authors Download PDF Abstract: AI-based protein structure prediction pipelines, such as AlphaFold2, haveĪchieved near-experimental accuracy.


 0 kommentar(er)
0 kommentar(er)
